deepwalk、node2vec的高性能优化
转自知乎: https://zhuanlan.zhihu.com/p/348778146
原作者:马东什么
首先是graph的存储问题,看了很多实现node2vec或deepwalk的library,大都是以networkx或是类似的python class对象为graph的存储形式进行开发,首先我们需要知道目前单机版的deepwalk和node2vec的主要的性能瓶颈在random walk部分,因为底层的word2vec基本上都是直接调用的gensim的word2vec(gensim的word2vec基于c++开发,目前新版的gensim的word2vec的fast version使用了cython代替了原来的一些逻辑实现,个人感觉从word2vec层面再去做优化太复杂了,也很难比gensim做的更好,除此之外另一个思路就是用tf.keras或torch来实现word2vec,借用gpu的算力来试图击败gensim,然而看完这几篇,我心灰意冷了:
Word2vec on GPU slower than CPU · Issue #13048 · tensorflow/tensorflow[github.comDoes Gensim library support GPU acceleration?stackoverflow.com
graph通常以相互引用的内存指针的方式存储在内存中,这意味着每个节点在内存中都位于不同的位置,这种基于基于链表的思想,使得内存访问的速度成为了主要的瓶颈,因为每次从一个节点移动到另一个节点,都需要在内存中寻址和查询,一种更好的方式是把节点都打包到数组中,因为数据的内存地址一般是连续的,在内存访问上的开销要小很多;(这里还没有提到networkx本身用纯python编写导致的较低效的编译效率问题,具体可见:
马东什么:为什么python这么慢?numba解决了什么问题?
)
一个较好的解决方案就是使用scipy.sparse.csr_matrix对graph进行存储和后续的访问等,
csr_matrix可见下:
马东什么:scipy sparse 中稀疏矩阵的常见存储方式zhuanlan.zhihu.com
可以看到,通过将graph转化为csr_matrix,利用其巧妙地存储方式,可以大大提高节点和节点之间的访问效率。
下面就来看一个非常nice的小众的graph library,csr_graph和nodevectors,nodevectors基本上是对csr_graph做的封装,底层的逻辑直接使用了csr_graph的接口api,所以直接研究csr_graph即可。
VHRanger/CSRGraphgithub.com
class csrgraph():
"""
This top level python class either calls external JIT'ed methods
or methods from the JIT'ed internal graph
"""
def __init__(self, data, nodenames=None, copy=True, threads=0):
"""
A class for larger graphs.
NOTE: this class tends to "steal data" by default.
If you pass a numpy array/scipy matrix/csr graph
Chances are this object will point to the same instance of data
Parameters:
-------------------
data : The graph data. Can be one of:
**NetworkX Graph**
**Numpy dense matrix**
**CSR Matrix**
**(data, indices, indptr)**
**CSRGraph object**
nodenames (array of str or int) : Node names
The position in this array should correspond with the node ID
So if passing a CSR Matrix or raw data, it should be co-indexed
with the matrix/raw data arrays
copy : bool
Wether to copy passed data to create new object
Default behavior is to point to underlying ctor passed data
For networkX graphs and numpy dense matrices we create a new object anyway
threads : int
number of threads to leverage for methods in this graph.
WARNING: changes the numba environment variable to do it.
Recompiles methods and changes it when changed.
0 is numba default (usually all threads)
TODO: add numpy mmap support for very large on-disk graphs
Should be in a different class
This also requires routines to read/write edgelists, etc. from disk
TODO: subclass scipy.csr_matrix???
"""
# If input is a CSRGraph, same object
if isinstance(data, csrgraph):
if copy:
self.mat = data.mat.copy()
self.names = deepcopy(data.names)
else:
self.mat = data.mat
self.names = data.names
if not nodenames:
nodenames = self.names
else:
self.names = nodenames
# NetworkX Graph input
elif isinstance(data, (nx.Graph, nx.DiGraph)):
mat = nx.adj_matrix(data)
mat.data = mat.data.astype(np.float32)
self.mat = mat
nodenames = list(data)
# CSR Matrix Input
elif isinstance(data, sparse.csr_matrix):
if copy: self.mat = data.copy()
else: self.mat = data
# Numpy Array Input
elif isinstance(data, np.ndarray):
self.mat = sparse.csr_matrix(data)
else:
raise ValueError(f"Incorrect input data type: {type(data).__name__}")
# Now that we have the core csr_matrix, alias underlying arrays
assert hasattr(self, 'mat')
self.weights = self.mat.data
self.src = self.mat.indptr
self.dst = self.mat.indices
# indptr has one more element than nnodes
self.nnodes = self.src.size - 1
# node name -> node ID
if nodenames is not None:
self.names = pd.Series(nodenames)
else:
self.names = pd.Series(np.arange(self.nnodes))
# Bounds check once here otherwise there be dragons later
max_idx = np.max(self.dst)
if self.nnodes < max_idx:
raise ValueError(f"""
Out of bounds node: {max_idx}, nnodes: {self.nnodes}
""")
self.set_threads(threads)
def set_threads(self, threads):
self.threads = threads
# Manage threading through Numba hack
if type(threads) is not int:
raise ValueError("Threads argument must be an int!")
if threads == 0:
threads = numba.config.NUMBA_DEFAULT_NUM_THREADS
threads = str(threads)
# If we change the number of threads, recompile
try:
prev_numba_value = os.environ['NUMBA_NUM_THREADS']
except KeyError:
prev_numba_value = threads
if threads != prev_numba_value:
os.environ['NUMBA_NUM_THREADS'] = threads
_random_walk.recompile()
_row_norm.recompile()
_node2vec_walks.recompile()
_node_degrees.recompile()
_src_multiply.recompile()
_dst_multiply.recompile()
def __getitem__(self, node):
"""
[] operator
like networkX, gets names of neighbor nodes
"""
# Get node ID from names array
# This is O(n) by design -- we more often get names from IDs
# than we get IDs from names and we don't want to hold 2 maps
# TODO : replace names with a pd.Index and use get_loc
node_id = self.names[self.names == node].index[0]
edges = self.dst[
self.src[node_id] : self.src[node_id+1]
]
return self.names.iloc[edges].values
def nodes(self):
"""
Returns the graph's nodes, in order
"""
if self.names is not None:
return self.names
else:
return np.arange(self.nnodes)
def normalize(self, return_self=True):
"""
Normalizes edge weights per node
For any node in the Graph, the new edges' weights will sum to 1
return_self : bool
whether to change the graph's values and return itself
this lets us call `G.normalize()` directly
"""
new_weights = _row_norm(self.weights, self.src)
if return_self:
self.mat = sparse.csr_matrix((new_weights, self.dst, self.src))
# Point objects to the correct places
self.weights = self.mat.data
self.src = self.mat.indptr
self.dst = self.mat.indices
gc.collect()
return self
else:
return csrgraph(sparse.csr_matrix(
(new_weights, self.dst, self.src)),
nodenames=self.names)
def random_walks(self,
walklen=10,
epochs=1,
start_nodes=None,
normalize_self=False,
return_weight=1.,
neighbor_weight=1.):
"""
Create random walks from the transition matrix of a graph
in CSR sparse format
Parameters
----------
T : scipy.sparse.csr matrix
Graph transition matrix in CSR sparse format
walklen : int
length of the random walks
epochs : int
number of times to start a walk from each nodes
return_weight : float in (0, inf]
Weight on the probability of returning to node coming from
Having this higher tends the walks to be
more like a Breadth-First Search.
Having this very high (> 2) makes search very local.
Equal to the inverse of p in the Node2Vec paper.
explore_weight : float in (0, inf]
Weight on the probability of visitng a neighbor node
to the one we're coming from in the random walk
Having this higher tends the walks to be
more like a Depth-First Search.
Having this very high makes search more outward.
Having this very low makes search very local.
Equal to the inverse of q in the Node2Vec paper.
threads : int
number of threads to use. 0 is full use
Returns
-------
out : 2d np.array (n_walks, walklen)
A matrix where each row is a random walk,
and each entry is the ID of the node
"""
# Make csr graph
if normalize_self:
self.normalize(return_self=True)
T = self
else:
T = self.normalize(return_self=False)
n_rows = T.nnodes
if start_nodes is None:
start_nodes = np.arange(n_rows)
sampling_nodes = np.tile(start_nodes, epochs)
# Node2Vec Biased walks if parameters specified
if (return_weight > 1. or return_weight < 1.
or neighbor_weight < 1. or neighbor_weight > 1.):
walks = _node2vec_walks(T.weights, T.src, T.dst,
sampling_nodes=sampling_nodes,
walklen=walklen,
return_weight=return_weight,
neighbor_weight=neighbor_weight)
# much faster implementation for regular walks
else:
walks = _random_walk(T.weights, T.src, T.dst,
sampling_nodes, walklen)
return walks
初始化init部分的逻辑很简单,就是把graph用scipy.sparse.csr_matrix的方式存储,需要注意的是,csr matrix仅支持有向/无向+有权/无权的简单图的表示,如果是复杂的属性图,例如存在多重的边关系或者是多个节点类型,则表示起来比较困难,不过常见的非GNN的graph embedding(以及排除了matapath2vec)之外大部分的graph embedding算法都是在简单图上运行的。
首先注意一个细节部分就是,我们要对边的权重做标准化norm,
new_weights = _row_norm(self.weights, self.src)
其实现逻辑为:
@jit(nopython=True, parallel=True, nogil=True, fastmath=True)
def _row_norm(weights, src):
"""
Returns the weights for normalized rows in a CSR Matrix.
Parameters
----------
weights : array[float]
The data array from a CSR Matrix.
For a scipy.csr_matrix, is accessed by M.data
src : array[int]
The index pointer array from a CSR Matrix.
For a scipy.csr_matrix, is accessed by M.indptr
----------------
returns : array[float32]
The normalized data array for the CSR Matrix
"""
n_nodes = src.size - 1
res = np.empty(weights.size, dtype=np.float32)
for i in numba.prange(n_nodes):
s1 = src[i]
s2 = src[i+1]
rowsum = np.sum(weights[s1:s2])
res[s1:s2] = weights[s1:s2] / rowsum
return res
可以看到,这里的逻辑就是把每一个节点的权重进行归一化,即每一个节点的和它连接的边的权重的权重之和为1,这里的实现就是每一行所有非零值初一这一行的非0值的总和。
可以看到,这里使用了numba和numba.prange进行了加速,需要注意的是,numba目前对于numpy的支持是最完善的,但是numba不支持scipy sparse!因此,我们将csr matrix的三要素:
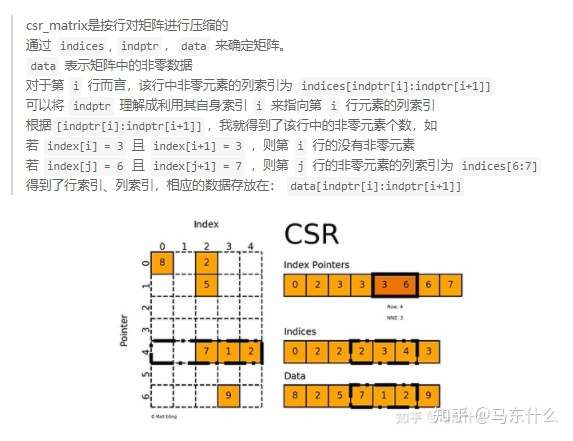
indices,indtpr,data用三个numpy array来存储,从而使用numba来间接优化scipy sparse,代码中的src是indtpr,
这里还是说明一下,如上图,index pointers即indptr,第i个节点具有的邻节点个数就是indptr[i+1]-indptr[1],第i个节点的邻节点的集合的列索引就是 indptr[i:i+1],例如对于节点4来说,indptr[4:5]=3:6=[3,4,5],然后去indices中查找,indices[3,4,5]=[2,3,4],此时我们就知道了 第四行的第2,3,4列(注意,初始都是从0开始的)是有值的,因此节点4的邻节点就是节点2和节点3(节点4和本身存在自环self loop),通过这种方式可以非常迅速地找到某个节点的邻节点,如果要找寻二阶邻节点,我们可以通过numba.prange再写一个循环,比如对于节点4的邻节点[2,3],我们循环对csr matrix矩阵的第二行和第三行进行同样的查询就可以了,访问非常的迅速,主要原因在于csr matrix本质上三个连续内存地址的数组,内存访问的效率很高。
对于numba,这里的装饰器设置:
@jit(nopython=True, parallel=True, nogil=True, fastmath=True)
比较固定,一般设置成这样就行,如果不涉及到并行则parallel可以不设置。
然后是核心的random walk部分:
def random_walks(self,
walklen=10,
epochs=1,
start_nodes=None,
normalize_self=False,
return_weight=1.,
neighbor_weight=1.):
"""
Create random walks from the transition matrix of a graph
in CSR sparse format
Parameters
----------
T : scipy.sparse.csr matrix
Graph transition matrix in CSR sparse format
walklen : int
length of the random walks
epochs : int
number of times to start a walk from each nodes
return_weight : float in (0, inf]
Weight on the probability of returning to node coming from
Having this higher tends the walks to be
more like a Breadth-First Search.
Having this very high (> 2) makes search very local.
Equal to the inverse of p in the Node2Vec paper.
explore_weight : float in (0, inf]
Weight on the probability of visitng a neighbor node
to the one we're coming from in the random walk
Having this higher tends the walks to be
more like a Depth-First Search.
Having this very high makes search more outward.
Having this very low makes search very local.
Equal to the inverse of q in the Node2Vec paper.
threads : int
number of threads to use. 0 is full use
Returns
-------
out : 2d np.array (n_walks, walklen)
A matrix where each row is a random walk,
and each entry is the ID of the node
"""
# Make csr graph
if normalize_self:
self.normalize(return_self=True)
T = self
else:
T = self.normalize(return_self=False)
n_rows = T.nnodes
if start_nodes is None:
start_nodes = np.arange(n_rows)
sampling_nodes = np.tile(start_nodes, epochs)
# Node2Vec Biased walks if parameters specified
if (return_weight > 1. or return_weight < 1.
or neighbor_weight < 1. or neighbor_weight > 1.):
walks = _node2vec_walks(T.weights, T.src, T.dst,
sampling_nodes=sampling_nodes,
walklen=walklen,
return_weight=return_weight,
neighbor_weight=neighbor_weight)
# much faster implementation for regular walks
else:
walks = _random_walk(T.weights, T.src, T.dst,
sampling_nodes, walklen)
return walks
可以看到,为了实现使用numba加速,基本上需要把所有的变量都以numpy形式存储,例如range使用np.arange,sampling_nodes = np.tile(start_nodes, epochs)等,numba目前也勉强支持list,dict这类的python数据结构,但是本身numpy的性能和扩展性都要好很多,而且也更加成熟,所以还是建议走numpy不容易各种bug。
最后就进入核心的核心了:
if (return_weight > 1. or return_weight < 1.
or neighbor_weight < 1. or neighbor_weight > 1.):
walks = _node2vec_walks(T.weights, T.src, T.dst,
sampling_nodes=sampling_nodes,
walklen=walklen,
return_weight=return_weight,
neighbor_weight=neighbor_weight)
# much faster implementation for regular walks
else:
walks = _random_walk(T.weights, T.src, T.dst,
sampling_nodes, walklen)
首先来看下_random_walk
@(nopython=True, parallel=True, nogil=True, fastmath=True)
def _random_walk(weights, indptr, dst,
sampling_nodes, walklen):
"""
Create random walks from the transition matrix of a graph
in CSR sparse format
Parameters
----------
weights : 1d np.array weights表示的是csr matrix的直接取值,使用csr matrix
存储的话,会有3个id array矩阵,分别是indices,indptr,data,含义在之前关于scipy的文章里
这里的weights对应的是data。
CSR data vector from a sparse matrix. Can be accessed by M.data
indptr : 1d np.array 这里的indptr对应的是csr matrix的indptr
CSR index pointer vector from a sparse matrix.
Can be accessed by M.indptr
dst : 1d np.array 这里的dst对应的是csr matrix的indices
CSR column vector from a sparse matrix.
Can be accessed by M.indices
sampling_nodes : 1d np.array of int samling nodes为我们要采样的节点,如果没有指定需要采样的
特定的节点则默认是对整个graph的所有节点进行采样
List of node IDs to start random walks from.
If doing sampling from walks, is equal to np.arrange(n_nodes)
repeated for each epoch
walklen : int random walks的长度,游走的次数在更上层的函数里,用epochs来表示,这里实际上是
length of the random walks
Returns
-------
out : 2d np.array (n_walks, walklen)
A matrix where each row is a random walk,
and each entry is the ID of the node
"""
n_walks = len(sampling_nodes) ##游走的次数
res = np.empty((n_walks, walklen), dtype=np.uint32) #用于存放游走的结果序列
n_nodes = indptr.size #节点数量
n_edges = weights.size #边数量
for i in numba.prange(n_walks):#对每个节点实施下面的逻辑,这里对每个节点的游走采样
#是完全独立可并行的,所以用numba的prange来自动实现对循环的并行,非常的直观方便
# Current node (each element is one walk's state)
state = sampling_nodes[i]
for k in range(walklen-1): #游走n次则进行n次循环,这里的循环时顺序执行的,所以无法并行
# Write state
res[i, k] = state # 初始节点,即start node
# Find row in csr indptr
start = indptr[state] #通过indptr来找到start node的邻节点
# 前面已经提到过csr matrix的indptr如何访问邻节点的问题了
end = indptr[state+1]
# If there are edges in the node, find next step
if start != end:#如果start!=end,这里用于判断是否无法继续游走(即游走到的节点没有邻
节点或者其邻节点在上一步已经游走过了),如果
#可以继续则继续执行下面的代码
# transition probabilities
p = weights[start:end] #根据indptr定位start node的邻节点,通过weights
#和indptr返回的列索引定位到start node和其邻节点之间的edges的权重的具体的值
# cumulative distribution of transition probabilities
cdf = np.cumsum(p)
# Random draw in [0, 1] for each row
# Choice is where random draw falls in cumulative distribution
draw = np.random.rand()
# Find where draw is in cdf
# Then use its index to update state
next_idx = np.searchsorted(cdf, draw) #上面通过简单的三个np函数就实现了
#随机采样,注意这里实现的是deepwalk的无偏简单采样,所以逻辑很简单,searchsorted在这个
#下非常合适,其作用
#在于在数组a中插入数组v(并不执行插入操作),返回一个下标列表,
#这个列表指明了v中对应元素应该插入在a中那个位置上,通过这种方式用numpy就实现了随机采样的简单逻辑了
# Winner points to the column index of the next node
state = dst[start + next_idx]#此时我们的start node发生改变,这里涉及到下一个节点的
#寻址,可以看到,我们通过dst,即indices的列表可以非常方便的定位到下一个节点的行位置。
# If there are no edges, this is the end of the walk
else:
res[i, k:] = state #存放采样结果
break
# Write final states
res[i, -1] = state#存放最终采样结果,即最后一个采样的节点
return res
真是越来越爱numba了。。。主要在于相对于cython来说,numba的使用更简单,你基本上用numpy的逻辑来写就行,对于numpy的死忠粉来说几乎无缝衔接。
这里的思路也不复杂,为了方便注释直接写代码里
下面i 看看node2vec的逻辑,原作者还没有实现rejective sampling有偏采样逻辑,具体在issues里面有提到过,不过这里有一个类似的版本中也是用numba实现了rejective sampling:
Accelerating node2vec with rejection samplinglouisabraham.github.io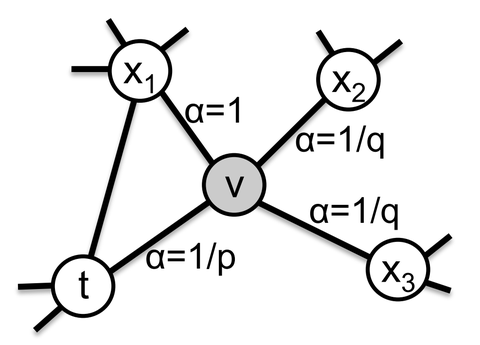 https://github.com/louisabraham/fastnode2vecgithub.com
https://github.com/louisabraham/fastnode2vecgithub.com
感觉这两个library可以做一个合并了。。。
import numpy as np
from numba import jit
@jit(nogil=True,nopython=True,parallel=True,fastmath=True)
def random_walk(walk_length, p, q, t):
"""sample a random walk starting from t
"""
# Normalize the weights to compute rejection probabilities
max_prob = max(1 / p, 1, 1 / q)
prob_0 = 1 / p / max_prob
prob_1 = 1 / max_prob
prob_2 = 1 / q / max_prob
# Initialize the walk
walk = np.empty(walk_length, dtype=indices.dtype)
walk[0] = t
walk[1] = random_neighbor(t)
for j in numba.prange(2, walk_length):
while True:
new_node = random_neighbor(walk[j - 1])
r = np.random.rand()
if new_node == walk[j - 2]:
# back to the previous node
if r < prob_0:
break
elif is_neighbor(walk[j - 2], new_node):
# distance 1
if r < prob_1:
break
elif r < prob_2:
# distance 2
break
walk[j] = new_node
return walk
我们看一下node2vec的有偏采样的逻辑,csrgraph怎么写的:
@jit(nopython=True, nogil=True, parallel=True, fastmath=True)
def _node2vec_walks(Tdata, Tindptr, Tindices,
sampling_nodes,
walklen,
return_weight,
neighbor_weight):
"""
Create biased random walks from the transition matrix of a graph
in CSR sparse format. Bias method comes from Node2Vec paper.
Parameters
----------
Tdata : 1d np.array casrmatrix的data
CSR data vector from a sparse matrix. Can be accessed by M.data
Tindptr : 1d np.array casrmatrix的indptr
CSR index pointer vector from a sparse matrix.
Can be accessed by M.indptr
Tindices : 1d np.array casrmatrix的indices
CSR column vector from a sparse matrix.
Can be accessed by M.indices
sampling_nodes : 1d np.array of int 采样节点,默认是全部节点
List of node IDs to start random walks from.
Is generally equal to np.arrange(n_nodes) repeated for each epoch
walklen : int 随机游走的步数
length of the random walks
return_weight : float in (0, inf] 返回权重,对应node2vec论文中的p
Weight on the probability of returning to node coming from
Having this higher tends the walks to be
more like a Breadth-First Search.
Having this very high (> 2) makes search very local.
return weights如果大于2则游走非常接近局部
Equal to the inverse of p in the Node2Vec paper.
explore_weight : float in (0, inf] # 探索权重,对应node2vec论文中的q
Weight on the probability of visitng a neighbor node
to the one we're coming from in the random walk
Having this higher tends the walks to be
more like a Depth-First Search.
Having this very high makes search more outward.
Having this very low makes search very local.
Equal to the inverse of q in the Node2Vec paper.
Returns
-------
out : 2d np.array (n_walks, walklen)
A matrix where each row is a biased random walk,
and each entry is the ID of the node
"""
n_walks = len(sampling_nodes)
res = np.empty((n_walks, walklen), dtype=np.uint32)
for i in numba.prange(n_walks):
# Current node (each element is one walk's state)
state = sampling_nodes[i]
res[i, 0] = state
# Do one normal step first
state = _node2vec_first_step(state, Tdata, Tindices, Tindptr)
for k in range(1, walklen-1):
# Write state
res[i, k] = state
state = _node2vec_inner(
res, i, k, state,
Tdata, Tindices, Tindptr,
return_weight, neighbor_weight
)
# Write final states
res[i, -1] = state
return res
这里实现了两个函数,一个是_node2vec_first_step用于有偏游走的初始化,一个是_node2vec_inner用于有偏游走后续的实现
其中:
@jit(nopython=True, nogil=True, fastmath=True)
def _node2vec_first_step(state, Tdata, Tindices, Tindptr):
"""
Inner code for node2vec walks
Normal random walk step
Comments for this logic are in _random_walk
"""
start = Tindptr[state]
end = Tindptr[state+1]
p = Tdata[start:end]
cdf = np.cumsum(p)
draw = np.random.rand()
next_idx = np.searchsorted(cdf, draw)
state = Tindices[start + next_idx]
return state
这里和deepwalk的逻辑是一样的,实际上有偏的定义是从初始游走之后的第一个游走才开始的,这一点建议对照论文的图仔细看看:
马东什么:Node2vec 深入分析](https://zhuanlan.zhihu.com/p/344839298)
也就是初始先随机走一步,然后开始下面正式的有偏游走:
@jit(nopython=True, nogil=True, fastmath=True)
def _node2vec_inner(
res, i, k, state,
Tdata, Tindices, Tindptr,
return_weight, neighbor_weight):
"""
Inner loop core for node2vec walks
Does the biased walk updating (pure function)
All arguments are directly from the node2vec walks method
"""
# Find rows in csr indptr
prev = res[i, k-1]
start = Tindptr[state] #当前节点的start和end,即得到当前节点的邻节点个数
end = Tindptr[state+1]
start_prev = Tindptr[prev] #当前节点的上一个节点(即从上一个节点游走到当前节点的
#上一个节点)的start和end,即得到上一个节点的邻节点个数
end_prev = Tindptr[prev+1]
# Find overlaps and fix weights
this_edges = Tindices[start:end] #当前节点的邻节点确定
prev_edges = Tindices[start_prev:end_prev] #上一个节点的邻节点确定
p = np.copy(Tdata[start:end]) #当前节点和邻节点连接的edges的权重
ret_idx = np.where(this_edges == prev) #确认上一个节点的index
p[ret_idx] = np.multiply(p[ret_idx], return_weight) #计算上一个节点
#和当前节点连接的edge权重*return weight
for pe in prev_edges:
n_idx_v = np.where(this_edges == pe)
n_idx = n_idx_v[0]
p[n_idx] = np.multiply(p[n_idx], neighbor_weight) #计算深入游走的概率和对应edge的积
# Get next state
## 剩下的部分就和deep walk一样了
cdf = np.cumsum(np.divide(p, np.sum(p)))
draw = np.random.rand()
next_idx = np.searchsorted(cdf, draw)
new_state = this_edges[next_idx]
return new_state
这里 思路其实是把p和q的参数和edges的权重进行相乘然后转化为常规的deepwalk的决策过程:
cdf = np.cumsum(np.divide(p, np.sum(p)))
draw = np.random.rand()
next_idx = np.searchsorted(cdf, draw)
学好numpy,做一些性能不错的开发工作还是很ok的~一些不是很复杂的需求,用numpy+numba或者numpy+numba或者numpy+numba+cython的性能并不会比c++差。